Post-treatment¶
When the simulation, one needs to post-treat some outputs.
1D outputs¶
The previously generated 1D csv files can be post-treated by an external tool PyTolosa.
One can execute the following command line by replacing /your/path/to/working/directory by your working directory path :
Note
One can also post-treat 1D and 2D results all at once. We will present this method in Browsing for outputs.
pytolosa -wdir /your/path/to/working/directory/res/savefield_1d -ssh -u -v -c1D -del
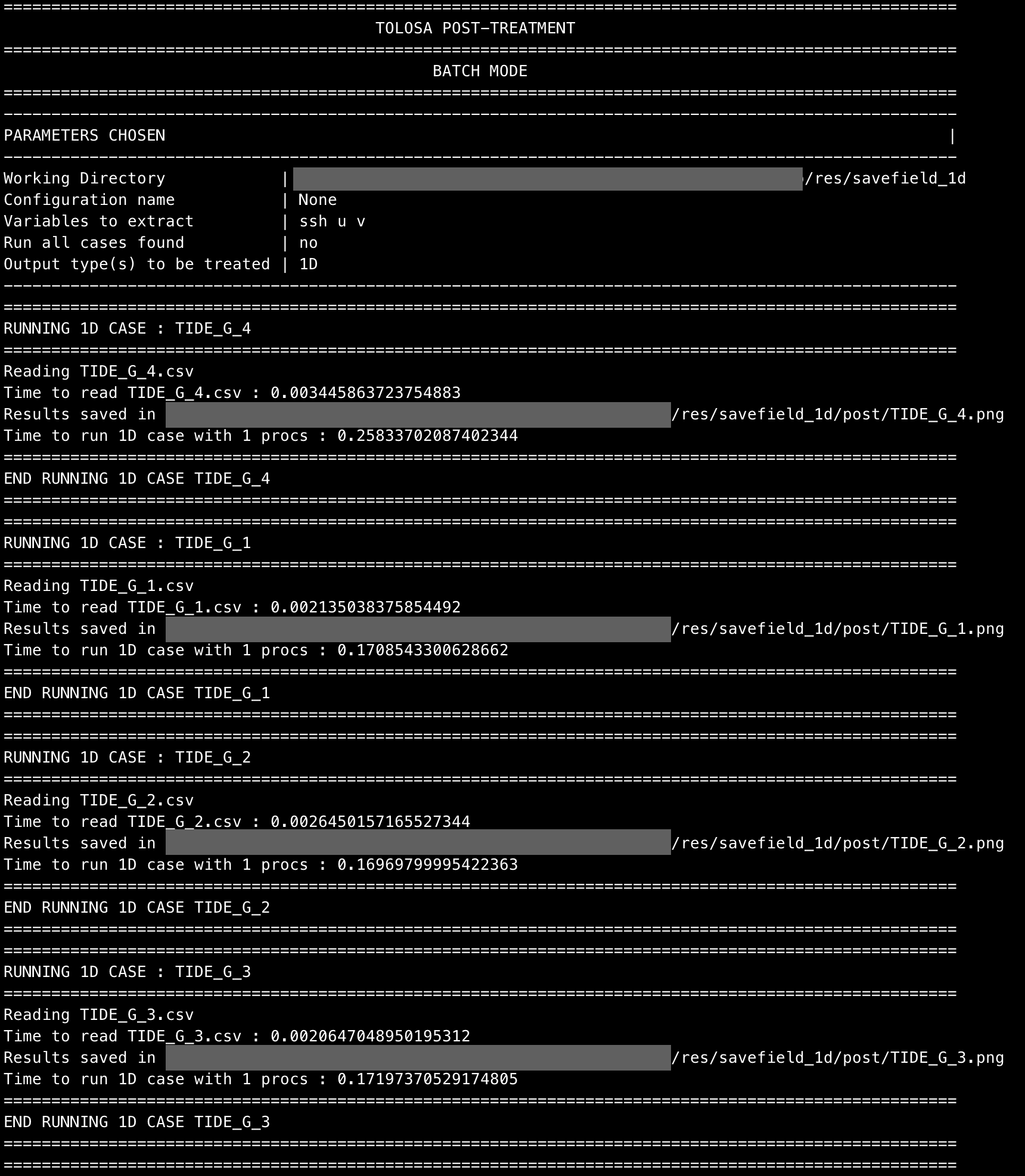
Four post-treatment images for each tide gauge should have been created in ./res/savefield_1d/post.
[IMAGES]
2D outputs¶
Each 2D output file has a different post-treatment method.
VTK Files¶
The previously generated VTK Files in ./res/vtk or ./res/savefield_2d/Window_1 can be read by Paraview. One can open the VTK files bundle :
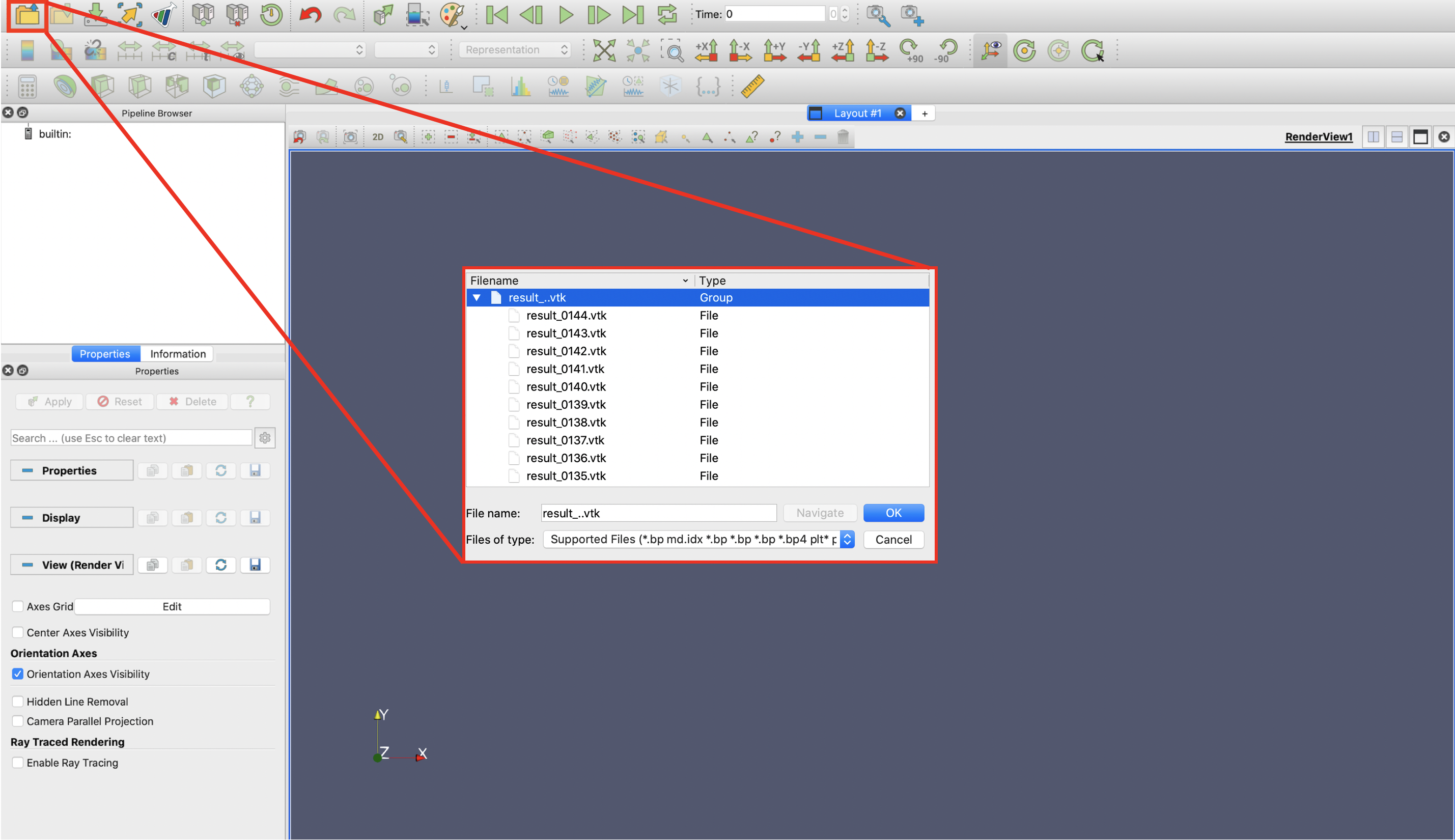
[RESULTS IMAGE]
PLT Files¶
PLT Files contain post-treatment data. One can display the results by using Gnuplot.
Binary Files¶
We also previously generated binary files. These files can be post-treated with PyTolosa.
One can execute the following command line by replacing /your/path/to/working/directory by your working directory path :
pytolosa -wdir /your/path/to/working/directory/res/savefield_2d -ssh -u -v -c2D -del -conf basic_river
One can also replace /res/savefield_2d/Window_2 if multiple windows with binary files were generated, and only the Window_2 needs to be post-treated.
[IMAGES]
Browsing for outputs - PyTolosa¶
PyTolosa enables the post-treatment of all 1D .csv and 2D binary outputs. This tool will explore all the ./res directory and search for files to be post-treated.
Therefore, one can execute the following command line to post-treat all output files :
pytolosa -wdir /your/path/to/working/director/res -ssh -u -v -c2D -del -conf basic_river -all
Note
Go to PyTolosa to learn more about all the execution options.