PyTolosa¶
Presentation¶
Some results generated by Tolosa-sw need to be post-treated. One could generate binary files, csv files with Tolosa, and post-treat them with PyTolosa. The different outputs generated by Tolosa are listed in Outputs.
PyTolosa enables the user to post-treat two-dimensional data from binary files, and one-dimensional data from csv files. To know how to generate those files from Tolosa, see Tutorial / Outputs.
In PyTolosa, two running modes are available : interactive mode and batch mode. Those two modes will be presented in Run.
System requirements¶
Environment¶
The following environment variable needs to be added to your ~/.bash_profile, or before executing PyTolosa, write :
export QT_QPA_PLATFORM='offscreen'
Python packages¶
The Python libraries and packages needed to use PyTolosa are :
- Matplotlib : creates static, animated, and interactive visualizations in Python.
- Numpy : fundamental package for scientific computing in Python.
- SciPy : collection of open source software for scientific computing in Python.
- CSV : implements classes to read and write tabular data in CSV format.
- mpi4py : provides bindings of the Message Passing Interface (MPI) standard, allowing any Python program to exploit multiple processors.
- argparse : writes user-friendly command-line interfaces.
- termcolor : ANSII Color formatting for output in terminal
One should also install FFMPEG to create animations of post-treated results.
Downloading¶
First, one must download the git repository to get the post-treatment program.
cd 'your_py_dir'
git clone https://github.com/TolosaProject/PyTolosa.git
Then, to be able to run PyTolosa and use its modules anywhere, add the path to the package in the PYTHONPATH environment variable, which will augment the default module search paths used by the Python interpreter.
Add the following line to your ~/.bashrc (or ~/.zshrc) to add permanently the environment variable and alias :
PYTHONPATH
Your path should end with /PyTolosa, where the pytolosa package is located.
If you do not wish to add permanently this PYTHONPATH and to create this alias, execute the following command line export PYTHONPATH=$PYTHONPATH:/your/path/to/PyTolosa, or place yourself in the /PyTolosa directory to run this post-treatment tool.
Principle and features¶
PyTolosa is a python package that enables to post-treat one dimensional and two dimensional Tolosa generated data. PyTolosa is able to read these 1D and 2D datas from .csv and .bin files respectively. It uses the matplotlib library to creates various plots.
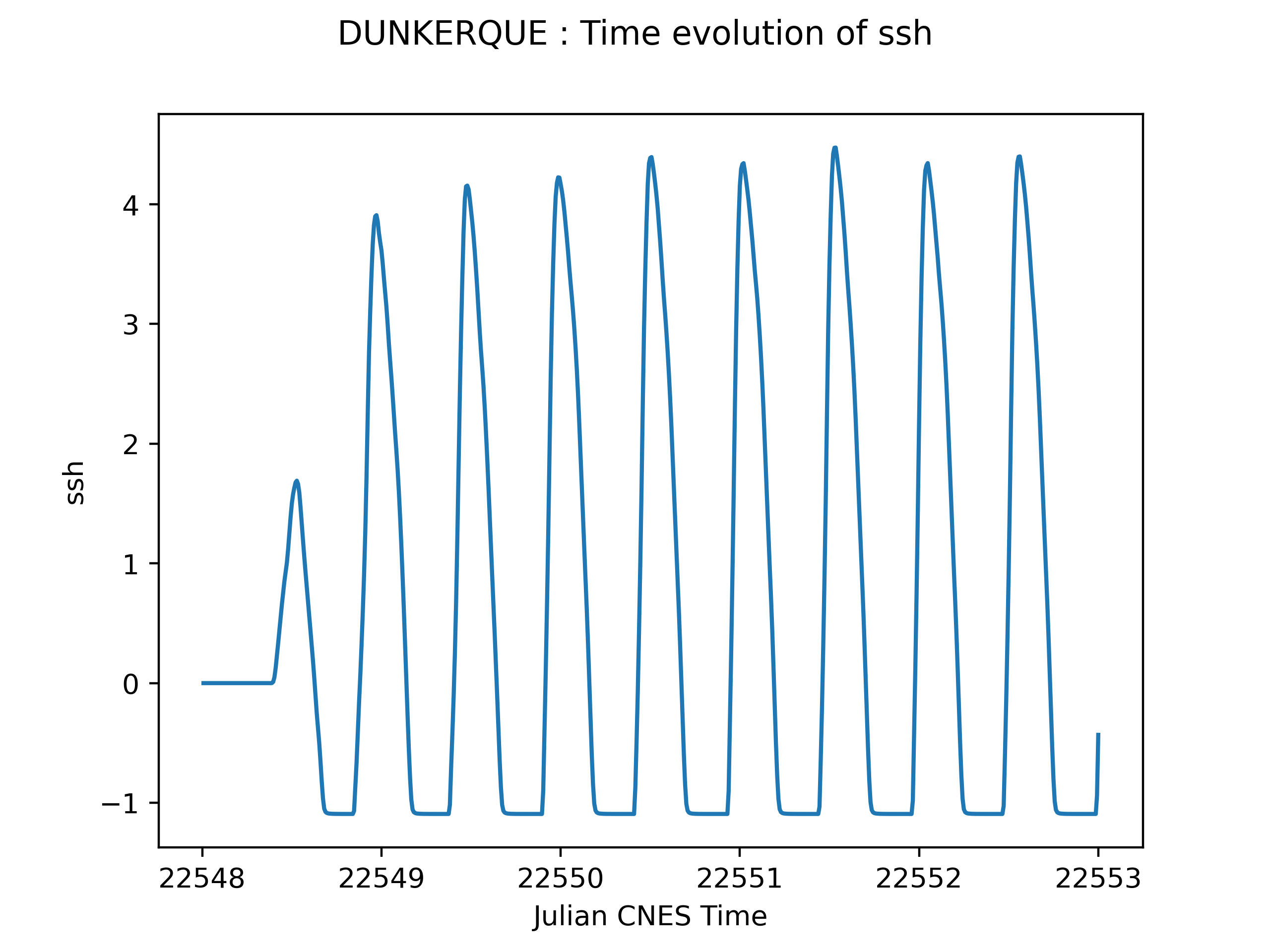
1D outputs¶
The one dimensional data can be post-treated if it is stored in a .csv file. PyTolosa reads those files and creates plots catching the time evolution of the fields extracted.
2D outputs¶
The two dimensional data can be post-treated if it is stored in .bin files. These files each represent the state of the domain at each output time step. One binary file contains :
- a key to define the content of the binary file.
Key signification
key = 0: No date and minimum and maximum values written at the beginning of the filekey = 1: The date is not written, but for each field, the minimum and maximum values are writtenkey = 2: The date is written, but no minimum and maximum valueskey = 3: The date and the minimum and maximum fields' values are written
- the date (if
key = 2or3) : written in seconds, gregorian date, and Julian CNES date. - the minimum and maximum values of the fields extracted (if
key = 1or3). - the fields values at each cell center. The fields that can be extracted (and read by PyTolosa) are the sea surface height, the global height, the bathymetry, and the vertical and horizontal components of the speed.
PyTolosa post-treat those files by creating an animation of the fields distribution on the domain. Thereby, PyTolosa needs to read an other binary file containing the mesh connectivity. By reading this mesh file and the other binary files (one for each time step), PyTolosa creates an animation in the directory containing those files.
Other features ...
- Change the display window : zoom on a certain subdomain by specifying the window (in longitude / latitude coordinates or in stereo x / y coordinates)
- Rescale the field's minimum and maximum display values : rescale the colorbar and define the new field's scale
- Animation settings : change the animation extension, the frame rate, the image size, the color of the colormap, and the image resolution
Browsing¶
PyTolosa also enables the user to explore his working directory to search the 1D and/or 2D cases found, and post-treat them automatically. One could activate this option by adding -all when running PyTolosa. All the options will be detailed in the Run section.
Run¶
Two running modes are available : an interactive mode and a batch mode.
Running informations
To get the running informations, write :
python3 -m pytolosa -h
Interactive mode¶
The interactive mode enables the user to enter all needed informations without listing the arguments himself (cf. Batch mode). The user will interact with the program to enter needed informations. The command lines to execute the program in this mode are the following :
- Serial
pytolosa -i
Without the PYTHONPATH environment variable or alias
Place yourself in the /PyTolosa directory.
python3 -m pytolosa -i
- Parallel
Run this post-treatment tool in parallel with 4 MPI processes :
mpirun -np 4 python3 -m pytolosa -i
Without the PYTHONPATH environment variable or alias
Place yourself in the /PyTolosa directory and run the same command.
Batch mode¶
The batch running mode enables the user to run this post-treatment program without any interaction. The existing running arguments are listed below.
- List of general arguments in non interactive mode :
| Arguments | Description |
|---|---|
-wdir WDIR |
Working Directory (./bin/res/...) with output directories |
-c1D |
Run 1D cases |
-c2D |
Run 2D cases |
-tmin TMIN |
Set the time to begin the data extraction. Format : Gregorian (YYYY-MM-DD_HH:MM) or Julian CNES date. |
-tmax TMAX |
Set the time to stop the data extraction. Format : Gregorian (YYYY-MM-DD_HH:MM) or Julian CNES date. |
-all |
Run all 1D and/or 2D cases found in directory tree |
-del, --delete |
Clean previous results in working directory |
-title TITLE |
Set title for results figures |
-ns, --nostream |
Access binary file without stream |
-verbose VERBOSE |
Verbosity level. From 0 to 2. |
- List of fields that could be extracted. Choose at least one field.
| Arguments | Description |
|---|---|
-h |
Reads h fields |
-ssh |
Reads ssh fields |
-u |
Reads u fields |
-v |
Reads v fields |
-bathy |
Reads bathymetry |
- 2D : arguments for 2D case(s).
| Arguments | Description |
|---|---|
-hmin HMIN |
Minimum height value considered |
-hmax HMAX |
Maximum height value considered |
-sshmin SSHMIN |
Minimum ssh value considered |
-sshmax SSHMAX |
Maximum ssh value considered |
-umin UMIN |
Minimum horizontal velocity value considered |
-umax UMAX |
Maximum horizontal velocity value considered |
-vmin VMIN |
Minimum vertical velocity value considered |
-vmax VMAX |
Maximum vertical velocity value considered |
-bmin BATHYMIN, --bathymin BATHYMIN |
Minimum bathymetry value considered |
-bmax BATHYMAX, --bathymax BATHYMAX |
Maximum bathymetry value considered |
-plottype PLOTTYPE |
Plot type if a smaller window is considered : static or dynamic. |
-xmin XMIN |
Set smaller display window with minimum x coordinate xmin |
-xmax XMAX |
Set smaller display window with maximum x coordinate xmax |
-ymin YMIN |
Set smaller display window with minimum y coordinate ymin |
-ymax YMAX |
Set smaller display window with maximum y coordinate ymax |
-lonmin LONGITUDEMIN, --longitudemin LONGITUDEMIN |
Set smaller display window with minimum longitude coordinate lonmin |
-lonmax LONGITUDEMAX, --longitudemax LONGITUDEMAX |
Set smaller display window with maximum longitude coordinate lonmax |
-latmin LATITUDEMIN, --latitudemin LATITUDEMIN |
Set smaller display window with minimum latitude coordinate latmin |
-latmax LATITUDEMAX, --latitudemax LATITUDEMAX |
Set smaller display window with maximum latitude coordinate latmax |
-an_fps AN_FPS |
Animation : frames per second |
-an_ext AN_EXT |
Animation : extension (def=mp4) |
-an_size AN_SIZE |
Animation : image size (give number of horizontal pixels) |
-cmap CMAP |
Colormap: new color range. See https://matplotlib.org/stable/gallery/color/colormap_reference.html |
-dpi DPI |
Image resolution in dots per inch. Default to 400. |
Info
Some arguments are mandatory, such as : -wdir, -c1D or -c2D (at least one case type), -h or -ssh or -u or -v or -bathy (at least one field).
The command lines to run PyTolosa in batch mode are the following :
- Serial
pytolosa [-args]
Without the PYTHONPATH environment variable or alias
Place yourself in the /PyTolosa directory and run the following command:
python3 -m pytolosa [-args]
- Parallel
Run this post-treatment tool in parallel with 4 MPI processes :
Without the PYTHONPATH environment variable or alias
Place yourself in the /PyTolosa directory and run the same command.